P01
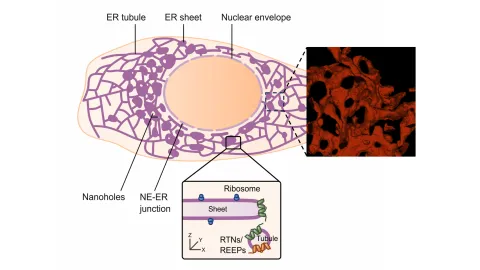
Project P01 characterizes the unique environments and functions of the different ER-membrane shaping Reticulons and REEPs.
Anne Schlaitz
P02
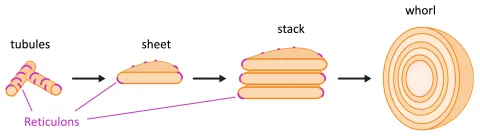
This project investigates the mechanisms of endoplasmic reticulum (ER) morphogenesis. Using yeast, we will examine the regulation of reticulon proteins and explore how ER stacks and whorls are formed.
Sebastian Schuck
P03
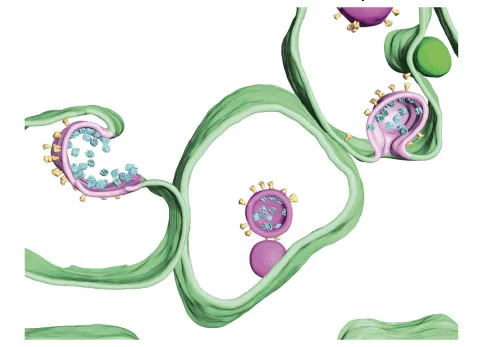
In situ cryo-ET of SARS-CoV-2 assembly
Petr Chlanda, Ulrich Schwarz
P04
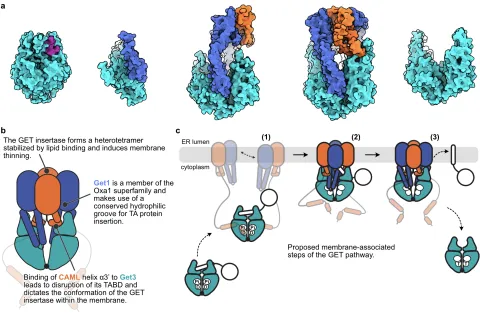
This project investigates the structural and functional implications of lipid-protein interactions for TA protein insertion and membrane morphology at the ER membrane.
Irmgard Sinning
P05
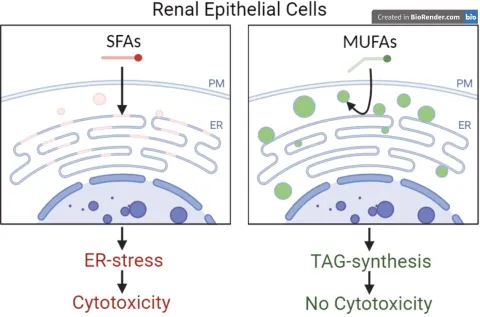
The uptake of albumin-bound fatty acids can cause lipotoxicity in renal proximal tubular cells. Here, we study the role of the E3 ubiquitin ligase RNF145 in this process by focussing on ER membrane fluidity and lipid droplet formation.
Matias Simons
P06
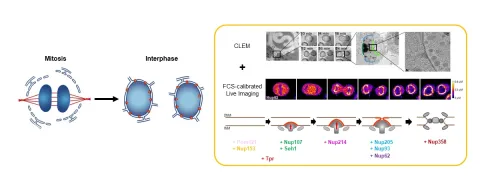
We study how nuclear membranes bend during NPC insertion, combining acute molecular perturbations with advanced imaging and lipidomics, to reveal principles of nuclear integrity, disease and evolution.
Jan Ellenberg
P07
The assembly of nuclear pore complexes (NPCs) during interphase begins with the recruitment of nucleoporins to the nuclear side of the inner nuclear membrane.
Elmar Schiebel
P08
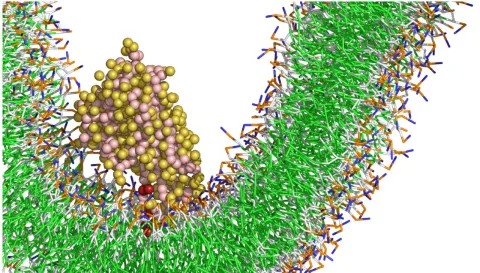
Exploring how mitochondrial membrane shape and composition control lipid transfer to uncover key principles of cellular organization and energy production.
Michael Meinecke
P09
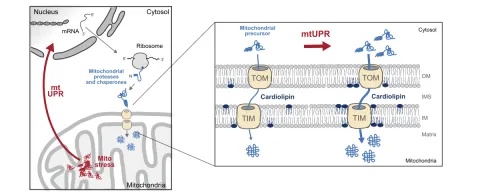
We investigate how mitochondrial stress triggers cardiolipin remodelling and lipid changes via the mtUPR to protect mitochondrial integrity and lipid homeostasis.
Nora Vögtle
P10
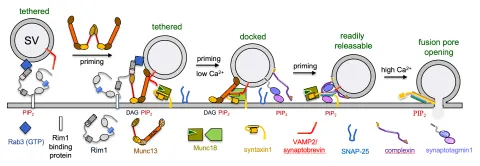
Project P10 will biochemically and structurally characterize the events corresponding to synaptic vesicle tethering at the active zone, involving Rab3 and its interacting partner Rim1.
Thomas Söllner
P11
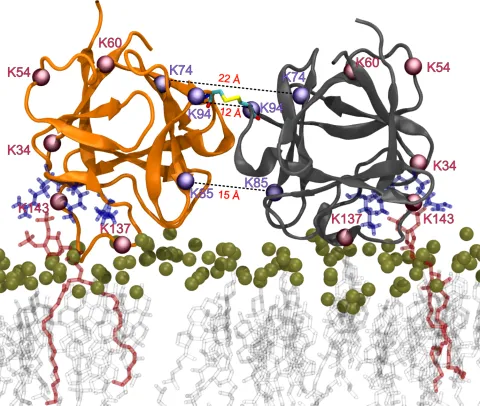
Disulfide bond formation enables the translocation of FGF2 across the plasma membrane. We aim to identify the source of oxidant and the mechanism of FGF2 oxidation.
Walter Nickel, Tobias Dick
P12
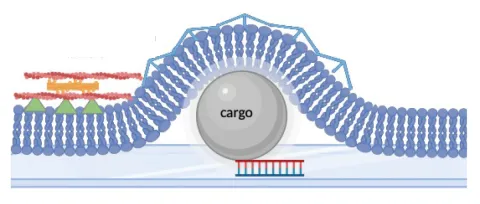
In this project we aim to understand how the mechanical properties of a cell, and in particular cell surface mechanics, influence membrane curvature during endocytosis.
Ada Cavalcanti-Adam, Alba Diz-Muñoz
P13
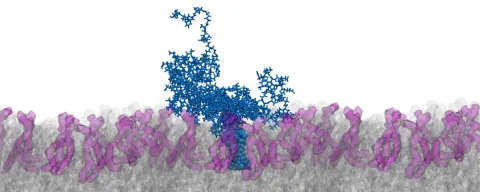
This project explores how membranes in their function as signaling platforms transmit mechanical stimuli into cells.
Frauke Gräter, Walter Nickel
P15
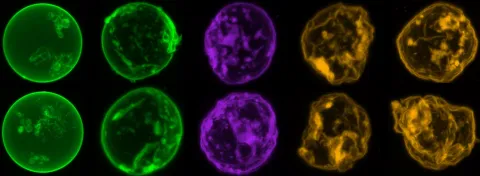
P15 engineers membrane-remodelling DNA and RNA nanostructures.
Kerstin Göpfrich
Z01

We use computer simulations to develop tools for setting up, and analyzing protein–membrane dynamics, providing insight into the complex process of membrane remodeling.
Frauke Gräter, Fabio Lolicato
Z02
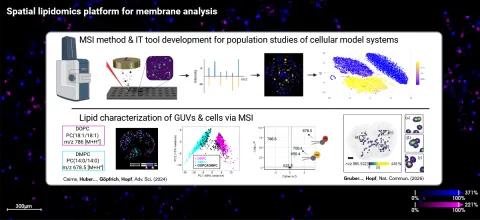
Our team develops new spatial and single cell lipidomics technologies for lipid remodelling research and applies them to support the CRC.
Britta Brügger, Carsten Hopf
Z03
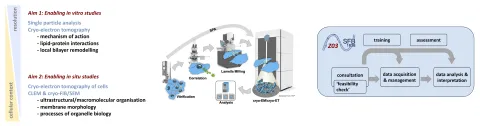
Development and application of Cryo-EM and cryo-CLEM methods for membrane biology research.
Petr Chlanda, Cristina Paulino
INF
The INF project supports the CRC research groups in transforming their scientific results into FAIR data.
Irmgard Sinning, Jürgen Kopp
